Thanks to the massive influx of newly resolved transmembrane protein structures (attributed to the revolution of cryo-EM and AlphaFold), selecting and visualizing a specific protein family grows harder by the day despite existing general and protein domain-specific databases. While the significant increase in the number of reliable structures for transmembrane proteins is a welcome change, the fact that the structures of a given family have to be collected from multiple data providers poses a potential challenge to performing comparative computational studies and to designing experiments.
In order to facilitate ABC protein related studies, we collected, classified, and exposed structures of ABC proteins via this web application.
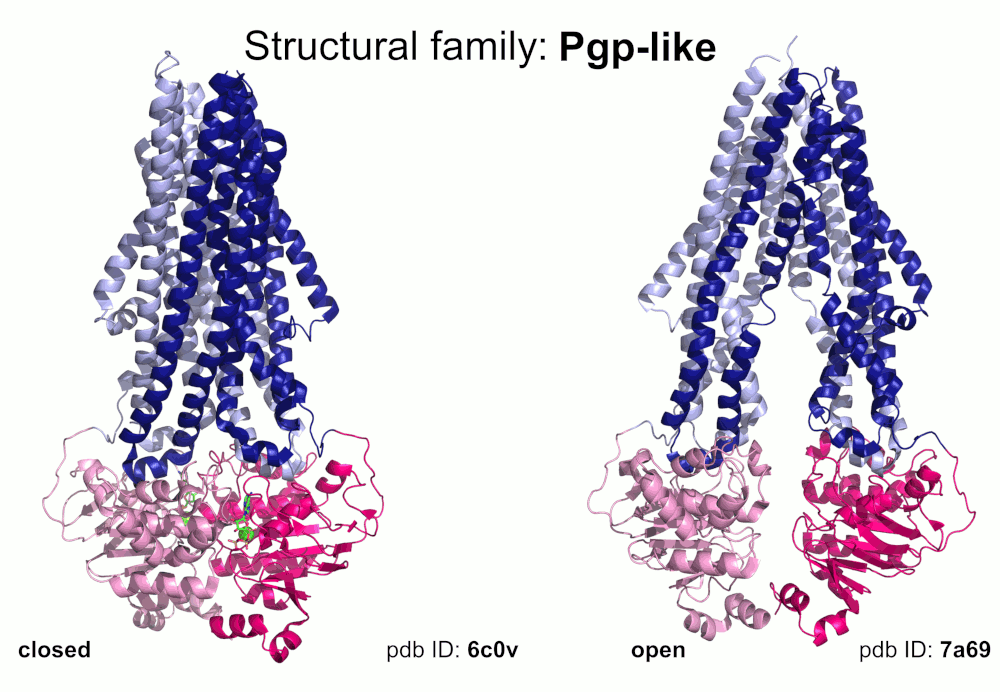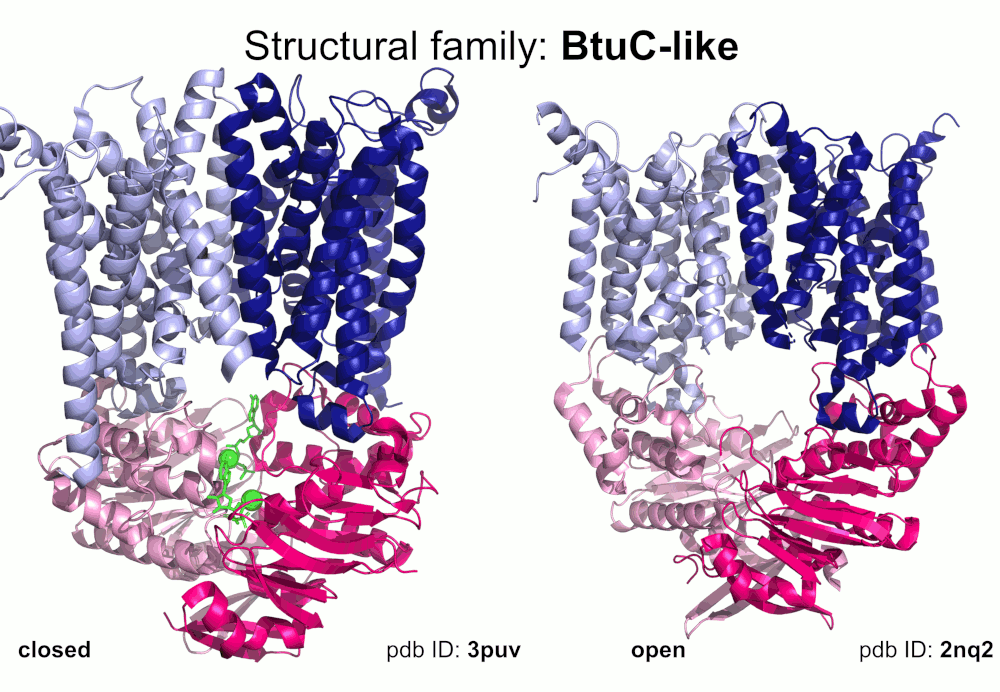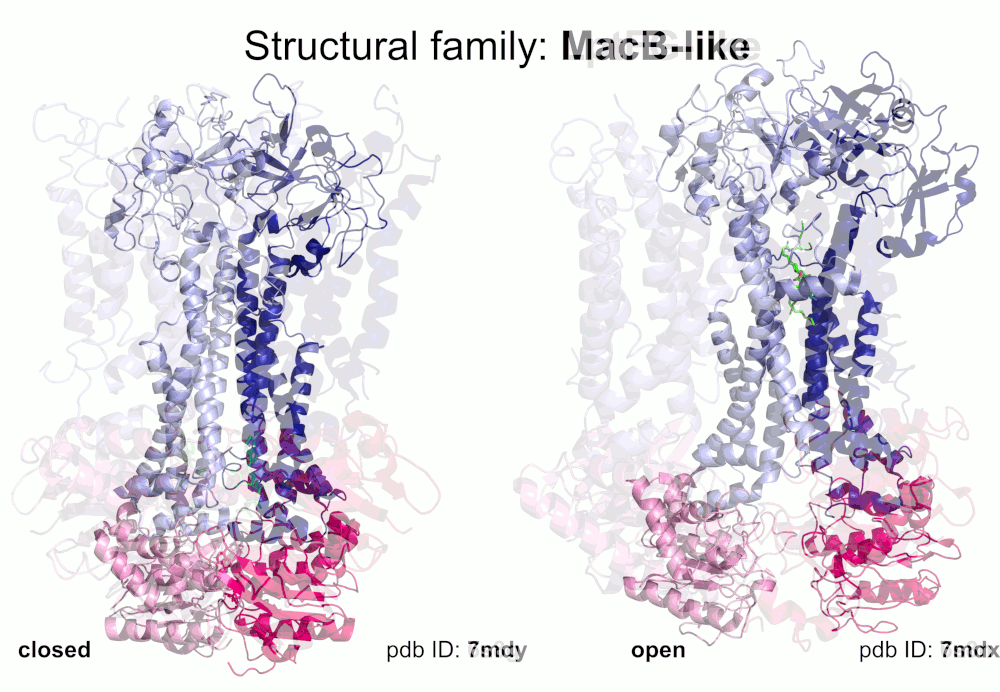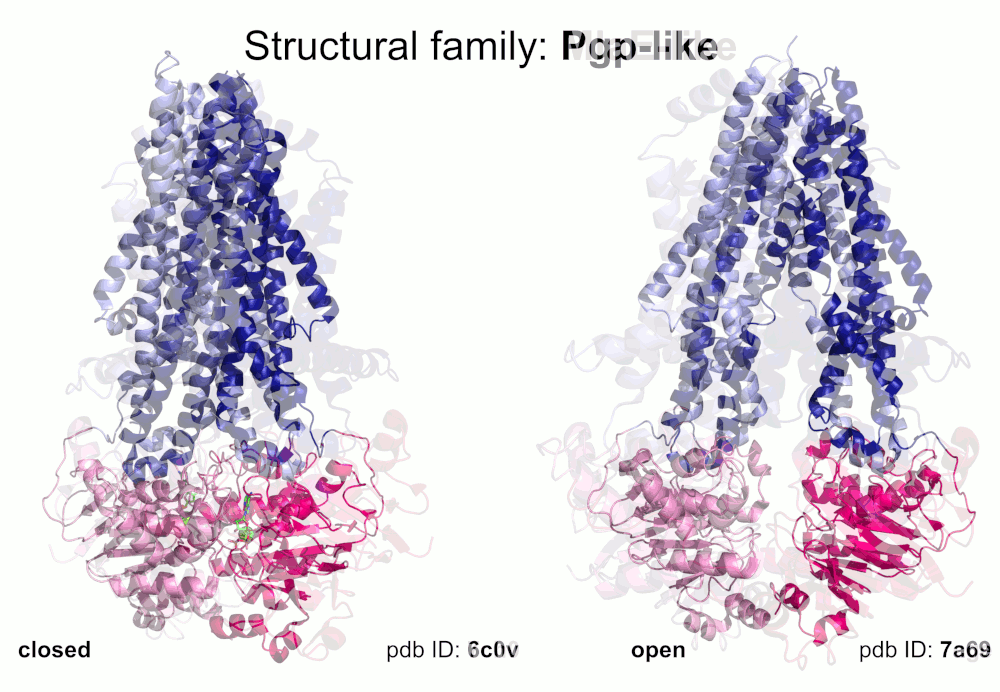
Our pipeline for ABC structure collection identifies and classifies transmembrane ABC protein structures using Pfam domain searches (the selected Pfam entries can be downloaded from here; we did not employ the DALI server, since it can not reliably distinguish different ABC TM folds - see here). Our classification is based on transmembrane domains, similar to that of Thomas et al.. However, instead of numbered structural classes we use names related to a well-known representative member of a class (e.g. Pgp-like and BtuCD-like). We find this approach not only more expressive than numbers, but the recently renumbered classes cause confusions in publications of the last decades (e.g. type I ABC exporter in the old system corresponds to type IV transporter structure in the new system). New structural folds (Bce-like and MlaE-like), which were determined after the publication of Thomas et al., were also inserted into our system. We also aimed to determine the conformational state of ABC structures based on a special geometric measure, the distance between the two nucleotide binding domains as length of the conftor(WA/SIG) (see our publication on conftors for Pgp-like structures).